In [8]:
mol = suppl[0]
mol
Out[8]:
import os
os.chdir('/public/gkxiao/work/dekois/vegfr2/vina')
加载库
from vina import Vina
import rdkit
from rdkit import Chem
from rdkit.Chem import AllChem,pdbqt
创建一个vina对象,并设置打分函数为vina
#create docking object
dock = Vina(sf_name='vina')
读入准备好的蛋白结构(Apo)
receptor = '3c7q_prot.pdbqt'
dock.set_receptor(receptor)
设置对接计算的盒子参数
# compute_vina_maps(self, center, box_size, spacing=0.375, force_even_voxels=False)
# Compute affinity maps using Vina scoring function.
center = [40.821, 25.760, 15.733]
box_size = [30, 30, 30]
dock.compute_vina_maps(center=center, box_size=box_size)
以ZINC08441965为小分子测试算例
import rdkit
from rdkit import Chem
from rdkit.Chem import AllChem
suppl = Chem.SDMolSupplier('ZINC08441965.sdf', removeHs=False)
加氢,距离几何法生成3D结构并进行能量优化
mol = suppl[0]
mol
title = mol.GetProp('_Name')
print(title)
ZINC08441965
将就化合物的名称添加为SDF的Tag
mol.SetProp('Title',title)
将对接的结果保存为sdf格式文件
output = Chem.SDWriter('ZINC08441965_debug_dock_20211225.sdf')
以输入的分子为模板,以便将对接的结果按模板归属键级
template = Chem.RemoveHs(mol)
开始对接计算
#将RDKIT分子转化为PDBQT
mol_pdbqt = pdbqt.MolToPDBQTBlock(mol, True, False, True)
#将PDBQT用于分子对接
dock.set_ligand_from_string(''.join(mol_pdbqt))
#设置穷尽数为32,给出9个pose
dock.dock(exhaustiveness=32, n_poses=9)
pose = dock.poses(n_poses=9, energy_range=5.0, coordinates_only=False)
poselines= pose.split('\n')
model_index=[]
end_index=[]
for i in range(len(poselines)):
if 'MODEL' in poselines[i]:
model_index.append(i)
if 'ENDMDL' in poselines[i]:
end_index.append(i)
for n in range(len(model_index)):
start_i = model_index[n]
end_i = end_index[n]
vina_pose = '\n'.join(poselines[start_i:end_i+1])
pose_rdkit = pdbqt.MolFromPDBQTBlock(vina_pose, sanitize=False, removeHs=True)
try:
pose_rdkit = Chem.RemoveHs(pose_rdkit)
#用模板归属键级
pose_corr = AllChem.AssignBondOrdersFromTemplate(template, pose_rdkit)
except:
smi_template = Chem.MolToSmiles(template)
smi_pose = Chem.MolToSmiles(pose_rdkit)
#如果键级归属失败,则输出模板与docking后识别的分子的SMILES格式
print("Failed molecule input:",smi_template,title)
print("Failed molecule pose:",smi_pose,title)
continue
pose_corr.SetProp('Title',title)
#pose_corr = Chem.AddHs(pose_corr)
output.write(pose_corr)
output.close()
Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965 Failed molecule input: [H]/N=C1\C(C#N)C2=CC[N@@H+](Cc3ccccc3)C[C@@H]2[C@@H](c2ccc(OC)c3ccccc23)C1(C#N)C#N ZINC08441965 Failed molecule pose: COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12 ZINC08441965
我们发现,这次键级归属失败,看看PDBQT转RDKit出现了什么问题,将2D结构呈现出来并与模板比较。先看看这次docking的pose。
print(vina_pose)
MODEL 9
REMARK VINA RESULT: -7.725 2.563 5.609
REMARK INTER + INTRA: -11.059
REMARK INTER: -9.758
REMARK INTRA: -1.301
REMARK UNBOUND: -1.301
REMARK Name = ZINC08441965
REMARK 5 active torsions:
REMARK status: ('A' for Active; 'I' for Inactive)
REMARK 1 A between atoms: _1 and _2
REMARK 2 A between atoms: _2 and _3
REMARK 3 A between atoms: _6 and _13
REMARK 4 A between atoms: _29 and _30
REMARK 5 A between atoms: _29 and _36
ROOT
ATOM 13 C12 UNL 1 41.189 31.873 21.493 0.00 0.00 0.034 C
ATOM 14 C13 UNL 1 42.508 31.214 21.902 0.00 0.00 0.041 C
ATOM 15 C14 UNL 1 42.860 31.505 23.343 0.00 0.00 0.085 C
ATOM 16 C15 UNL 1 43.224 29.172 23.829 0.00 0.00 0.096 C
ATOM 17 C16 UNL 1 42.762 28.804 22.440 0.00 0.00 -0.029 C
ATOM 18 C17 UNL 1 42.409 29.745 21.603 0.00 0.00 -0.036 C
ATOM 19 C18 UNL 1 41.842 29.358 20.287 0.00 0.00 0.108 C
ATOM 20 C19 UNL 1 41.179 30.287 19.551 0.00 0.00 0.065 C
ATOM 21 C20 UNL 1 41.001 31.712 19.984 0.00 0.00 0.189 C
ATOM 22 C21 UNL 1 39.646 32.160 19.620 0.00 0.00 0.091 C
ATOM 23 N1 UNL 1 38.601 32.505 19.339 0.00 0.00 -0.196 N
ATOM 24 C22 UNL 1 41.986 32.551 19.283 0.00 0.00 0.091 C
ATOM 25 N2 UNL 1 42.747 33.198 18.741 0.00 0.00 -0.196 N
ATOM 26 N3 UNL 1 40.645 29.902 18.353 0.00 0.00 -0.305 N
ATOM 27 C23 UNL 1 41.994 28.021 19.798 0.00 0.00 0.076 C
ATOM 28 N4 UNL 1 42.114 26.960 19.411 0.00 0.00 -0.197 N
ATOM 36 N5 UNL 1 42.482 30.388 24.223 0.00 0.00 -0.327 N
ATOM 46 H10 UNL 1 40.354 31.396 22.026 0.00 0.00 0.039 H
ATOM 47 H11 UNL 1 43.339 31.637 21.320 0.00 0.00 0.042 H
ATOM 48 H12 UNL 1 42.334 32.414 23.669 0.00 0.00 0.093 H
ATOM 49 H13 UNL 1 43.943 31.677 23.424 0.00 0.00 0.093 H
ATOM 50 H14 UNL 1 42.994 28.353 24.527 0.00 0.00 0.097 H
ATOM 51 H15 UNL 1 44.303 29.381 23.820 0.00 0.00 0.097 H
ATOM 52 H16 UNL 1 42.722 27.748 22.133 0.00 0.00 0.064 H
ATOM 53 H17 UNL 1 41.937 28.325 19.921 0.00 0.00 0.057 H
ATOM 54 H18 UNL 1 40.740 28.936 18.012 0.00 0.00 0.187 HD
ATOM 62 H26 UNL 1 41.471 30.211 24.133 0.00 0.00 0.348 HD
ENDROOT
BRANCH 13 6
ATOM 3 C2 UNL 1 41.335 36.061 22.472 0.00 0.00 0.126 A
ATOM 4 C3 UNL 1 42.322 35.469 21.736 0.00 0.00 -0.019 A
ATOM 5 C4 UNL 1 42.267 34.113 21.425 0.00 0.00 -0.054 A
ATOM 6 C5 UNL 1 41.229 33.338 21.842 0.00 0.00 -0.033 A
ATOM 7 C6 UNL 1 40.192 33.910 22.600 0.00 0.00 -0.011 A
ATOM 8 C7 UNL 1 40.247 35.287 22.926 0.00 0.00 0.024 A
ATOM 9 C8 UNL 1 39.209 35.860 23.678 0.00 0.00 -0.051 A
ATOM 10 C9 UNL 1 38.172 35.084 24.094 0.00 0.00 -0.061 A
ATOM 11 C10 UNL 1 38.120 33.726 23.781 0.00 0.00 -0.062 A
ATOM 12 C11 UNL 1 39.105 33.139 23.048 0.00 0.00 -0.054 A
ATOM 40 H4 UNL 1 43.174 36.071 21.386 0.00 0.00 0.066 H
ATOM 41 H5 UNL 1 43.075 33.661 20.830 0.00 0.00 0.063 H
ATOM 42 H6 UNL 1 39.235 36.930 23.931 0.00 0.00 0.063 H
ATOM 43 H7 UNL 1 37.362 35.534 24.688 0.00 0.00 0.062 H
ATOM 44 H8 UNL 1 37.271 33.121 24.130 0.00 0.00 0.062 H
ATOM 45 H9 UNL 1 39.051 32.067 22.808 0.00 0.00 0.063 H
BRANCH 3 2
ATOM 2 O1 UNL 1 41.404 37.385 22.768 0.00 0.00 -0.496 OA
BRANCH 2 1
ATOM 1 C1 UNL 1 42.423 38.143 22.114 0.00 0.00 0.078 C
ATOM 37 H1 UNL 1 42.297 37.969 21.036 0.00 0.00 0.067 H
ATOM 38 H2 UNL 1 43.382 37.698 22.419 0.00 0.00 0.067 H
ATOM 39 H3 UNL 1 42.379 39.215 22.360 0.00 0.00 0.067 H
ENDBRANCH 2 1
ENDBRANCH 3 2
ENDBRANCH 13 6
BRANCH 36 29
ATOM 29 C24 UNL 1 42.781 30.730 25.633 0.00 0.00 0.103 C
ATOM 55 H19 UNL 1 42.721 29.822 26.252 0.00 0.00 0.098 H
ATOM 56 H20 UNL 1 43.793 31.159 25.701 0.00 0.00 0.098 H
BRANCH 29 30
ATOM 30 C25 UNL 1 41.772 31.739 26.117 0.00 0.00 0.006 A
ATOM 31 C26 UNL 1 40.543 31.317 26.587 0.00 0.00 -0.053 A
ATOM 32 C27 UNL 1 39.619 32.242 27.036 0.00 0.00 -0.062 A
ATOM 33 C28 UNL 1 39.921 33.591 27.004 0.00 0.00 -0.062 A
ATOM 34 C29 UNL 1 41.148 34.014 26.530 0.00 0.00 -0.062 A
ATOM 35 C30 UNL 1 42.074 33.088 26.085 0.00 0.00 -0.053 A
ATOM 57 H21 UNL 1 40.301 30.244 26.605 0.00 0.00 0.063 H
ATOM 58 H22 UNL 1 38.644 31.905 27.418 0.00 0.00 0.062 H
ATOM 59 H23 UNL 1 39.184 34.327 27.358 0.00 0.00 0.062 H
ATOM 60 H24 UNL 1 41.388 35.087 26.505 0.00 0.00 0.062 H
ATOM 61 H25 UNL 1 43.051 33.424 25.707 0.00 0.00 0.063 H
ENDBRANCH 29 30
ENDBRANCH 36 29
TORSDOF 5
ENDMDL
把pose保存为pdbqt,以便用可视化软件观察。
dock.write_poses('ZINC08441965_debug_dock_20211225_out.pdbqt', n_poses=10, overwrite=True)
将其中一个pose转为smiles,并呈现它的2D图,你会发现在格式转化的过程中发生了键级错误:腈基变为氨基了,芳香环变为脂肪环。
rdkitmol = pdbqt.MolFromPDBQTBlock(vina_pose, sanitize=False, removeHs=True)
rdkitmol = Chem.RemoveHs(rdkitmol)
smi = Chem.MolToSmiles(rdkitmol)
print(smi)
mol_2D = Chem.MolFromSmiles(smi)
mol_2D
COC1CCC(C2C3C[NH+](CC4CCCCC4)CCC3C(CN)C(N)C2(CN)CN)C2CCCCC12
但是,原子的坐标与连接关系确是对的,因为你可以根据输出的PDBQT观察到,也可以从坐标文件观察到,如下所示:
print(Chem.MolToMolBlock(rdkitmol))
ZINC08441965
RDKit 3D
36 40 0 0 0 0 0 0 0 0999 V2000
42.4230 38.1430 22.1140 C 0 0 0 0 0 0 0 0 0 0 0 0
41.4040 37.3850 22.7680 O 0 0 0 0 0 0 0 0 0 0 0 0
41.3350 36.0610 22.4720 C 0 0 0 0 0 0 0 0 0 0 0 0
42.3220 35.4690 21.7360 C 0 0 0 0 0 0 0 0 0 0 0 0
42.2670 34.1130 21.4250 C 0 0 0 0 0 0 0 0 0 0 0 0
41.2290 33.3380 21.8420 C 0 0 0 0 0 0 0 0 0 0 0 0
40.1920 33.9100 22.6000 C 0 0 0 0 0 0 0 0 0 0 0 0
40.2470 35.2870 22.9260 C 0 0 0 0 0 0 0 0 0 0 0 0
39.2090 35.8600 23.6780 C 0 0 0 0 0 0 0 0 0 0 0 0
38.1720 35.0840 24.0940 C 0 0 0 0 0 0 0 0 0 0 0 0
38.1200 33.7260 23.7810 C 0 0 0 0 0 0 0 0 0 0 0 0
39.1050 33.1390 23.0480 C 0 0 0 0 0 0 0 0 0 0 0 0
41.1890 31.8730 21.4930 C 0 0 0 0 0 0 0 0 0 0 0 0
42.5080 31.2140 21.9020 C 0 0 0 0 0 0 0 0 0 0 0 0
42.8600 31.5050 23.3430 C 0 0 0 0 0 0 0 0 0 0 0 0
43.2240 29.1720 23.8290 C 0 0 0 0 0 0 0 0 0 0 0 0
42.7620 28.8040 22.4400 C 0 0 0 0 0 0 0 0 0 0 0 0
42.4090 29.7450 21.6030 C 0 0 0 0 0 0 0 0 0 0 0 0
41.8420 29.3580 20.2870 C 0 0 0 0 0 0 0 0 0 0 0 0
41.1790 30.2870 19.5510 C 0 0 0 0 0 0 0 0 0 0 0 0
41.0010 31.7120 19.9840 C 0 0 0 0 0 0 0 0 0 0 0 0
39.6460 32.1600 19.6200 C 0 0 0 0 0 0 0 0 0 0 0 0
38.6010 32.5050 19.3390 N 0 0 0 0 0 0 0 0 0 0 0 0
41.9860 32.5510 19.2830 C 0 0 0 0 0 0 0 0 0 0 0 0
42.7470 33.1980 18.7410 N 0 0 0 0 0 0 0 0 0 0 0 0
40.6450 29.9020 18.3530 N 0 0 0 0 0 0 0 0 0 0 0 0
41.9940 28.0210 19.7980 C 0 0 0 0 0 0 0 0 0 0 0 0
42.1140 26.9600 19.4110 N 0 0 0 0 0 0 0 0 0 0 0 0
42.7810 30.7300 25.6330 C 0 0 0 0 0 0 0 0 0 0 0 0
41.7720 31.7390 26.1170 C 0 0 0 0 0 0 0 0 0 0 0 0
40.5430 31.3170 26.5870 C 0 0 0 0 0 0 0 0 0 0 0 0
39.6190 32.2420 27.0360 C 0 0 0 0 0 0 0 0 0 0 0 0
39.9210 33.5910 27.0040 C 0 0 0 0 0 0 0 0 0 0 0 0
41.1480 34.0140 26.5300 C 0 0 0 0 0 0 0 0 0 0 0 0
42.0740 33.0880 26.0850 C 0 0 0 0 0 0 0 0 0 0 0 0
42.4820 30.3880 24.2230 N 0 0 0 0 0 0 0 0 0 0 0 0
14 13 1 0
15 14 1 0
17 16 1 0
18 17 1 0
18 14 1 0
19 18 1 0
20 19 1 0
21 20 1 0
21 13 1 0
22 21 1 0
23 22 1 0
24 21 1 0
25 24 1 0
26 20 1 0
27 19 1 0
28 27 1 0
36 16 1 0
36 15 1 0
4 3 1 0
5 4 1 0
6 13 1 0
6 5 1 0
7 6 1 0
8 7 1 0
8 3 1 0
9 8 1 0
10 9 1 0
11 10 1 0
12 11 1 0
12 7 1 0
2 3 1 0
1 2 1 0
29 36 1 0
30 29 1 0
31 30 1 0
32 31 1 0
33 32 1 0
34 33 1 0
35 30 1 0
35 34 1 0
M CHG 1 36 1
M END
现在,该OEChem登场了,利用OEChem的键级感知与电荷感知能力,将上面的坐标变为一个正确的分子。
isdf = Chem.MolToMolBlock(rdkitmol)+'$$$$'
import sys,os
sys.path.append('/public/gkxiao/software/OpenEye-toolkits-python3-linux-x64-2021.1.1')
from openeye import oechem
from openeye import oedepict
from openeye import oegrapheme
ifs = oechem.oemolistream()
ifs.SetFormat(oechem.OEFormat_SDF)
ifs.openstring(isdf)
True
mols = []
imol = oechem.OEMol()
for imol in ifs.GetOEMols():
print("Canonical isomeric SMILES before assigning bond orders is",oechem.OECreateCanSmiString(imol))
oechem.OEDetermineConnectivity(imol)
oechem.OEFindRingAtomsAndBonds(imol)
oechem.OEPerceiveBondOrders(imol)
oechem.OEAssignImplicitHydrogens(imol)
oechem.OEAssignFormalCharges(imol)
print("Canonical isomeric SMILES after assigning bond orders is", oechem.OEMolToSmiles(imol))
mols.append(oechem.OEMol(imol))
Canonical isomeric SMILES before assigning bond orders is COC1CCC(C2C1CCCC2)C3C4C[NH+](CCC4C(C(C3(CN)CN)N)CN)CC5CCCCC5 Canonical isomeric SMILES after assigning bond orders is COc1ccc(c2c1cccc2)[C@@H]3[C@H]4C[N@@H+](CC=C4C(=C(C3(C#N)C#N)N)C#N)Cc5ccccc5
画个图验证一下OEChem归属的键级与电荷是否正确,你可以将上面的第2行SMILES复制到ChemDraw上看看是否对。我用OEchem画这个结构,是这个样子。
oemol = mols[0]
oedepict.OEPrepareDepiction(oemol)
oedepict.OERenderMolecule("ZINC08441965-DepictMolSimple.png", oemol)
True
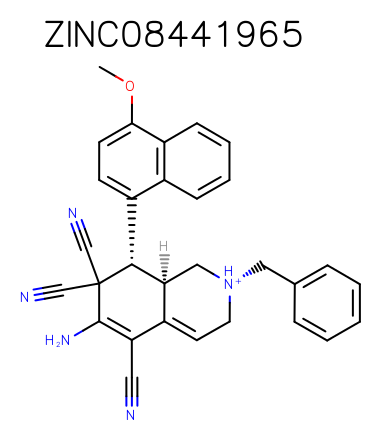
看起来,OEChem转化的得到分子与输入的起始结构完全一样,完美解决了键级与电荷的归属问题。