肖高铿 2021-12-28 http://blog.molcalx.com.cn
导入openeye软件包
import sys,os
sys.path.append('/public/gkxiao/software/OpenEye-toolkits-python3-linux-x64-2021.1.1')
from openeye import oechem
from openeye import oedepict
from openeye import oegrapheme
切换工作目录
os.chdir('/public/gkxiao/work/oechem_interaction/cdk6')
导入蛋白结构
ifile = '5l2s_prot.pdb'
ifs = oechem.oemolistream()
ifs.open(ifile)
prot = oechem.OEGraphMol()
oechem.OEReadMolecule(ifs, prot)
True
导入对接好的配体,本处从复合物结构里提取。
ifile = '5l2s_ligand.mol2'
ifs = oechem.oemolistream()
ifs.open(ifile)
Abemaciclib = oechem.OEGraphMol()
oechem.OEReadMolecule(ifs, Abemaciclib)
True
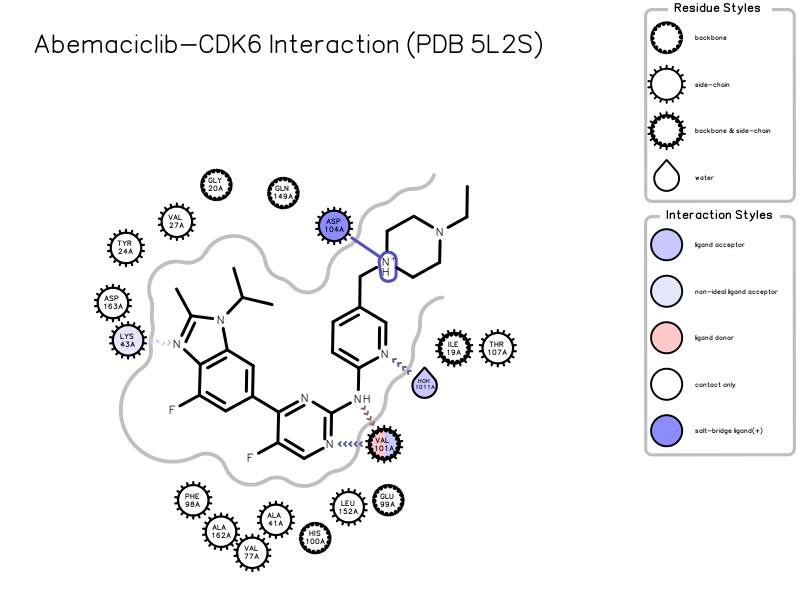
用OEChem进行相互作用分析
asite = oechem.OEInteractionHintContainer(prot, Abemaciclib)
oechem.OEPerceiveInteractionHints(asite)
True
用OEDipict进行绘图
oegrapheme.OEPrepareActiveSiteDepiction(asite)
True
设置图片的尺寸
asite.SetTitle('Abemaciclib-CDK6 Interaction (PDB 5L2S)')
width = 800
height = 600
image = oedepict.OEImage(width, height)
cframe = oedepict.OEImageFrame(image, width * 0.80, height, oedepict.OE2DPoint(0.0, 0.0))
lframe = oedepict.OEImageFrame(image, width * 0.20, height,
oedepict.OE2DPoint(width * 0.80, 0.0))
opts = oegrapheme.OE2DActiveSiteDisplayOptions(cframe.GetWidth(), cframe.GetHeight())
adisp = oegrapheme.OE2DActiveSiteDisplay(asite, opts)
oegrapheme.OERenderActiveSite(cframe, adisp)
lopts = oegrapheme.OE2DActiveSiteLegendDisplayOptions(12, 1)
oegrapheme.OEDrawActiveSiteLegend(lframe, adisp, lopts)
保存为图片:一张SVG格式,一张PNG格式
oedepict.OEWriteImage('5l2s-abemaciclib.svg', image)
oedepict.OEWriteImage('5l2s-abemaciclib.png', image)
True
