摘要:OEChem不仅是一个强大的数据整合工具,而且还是强大的蛋白-配体相互作用分析工具。本文以最近上市的ABL别构抑制剂Asciminib(ABL001)与ABL的复合物结构PDB 5MO4为例,演示了如何用oechem进行相互作用分析,以及自定义卤键相互作用的可选项,然后再用oedepict进行可视化,本文还演示了如何以文本的方式输出相互作用信息。
前言
OEChem不仅是一个强大的数据整合工具,而且还是强大的蛋白-配体相互作用分析工具。本文以最近上市的ABL别构抑制剂Asciminib(ABL001)与ABL的复合物结构PDB 5MO4为例,演示了如何用oechem进行相互作用分析,以及自定义卤键相互作用的参数,然后再用oedepict进行相互作用可视化。
操作步骤
下载生成的图片
下载生成的图片
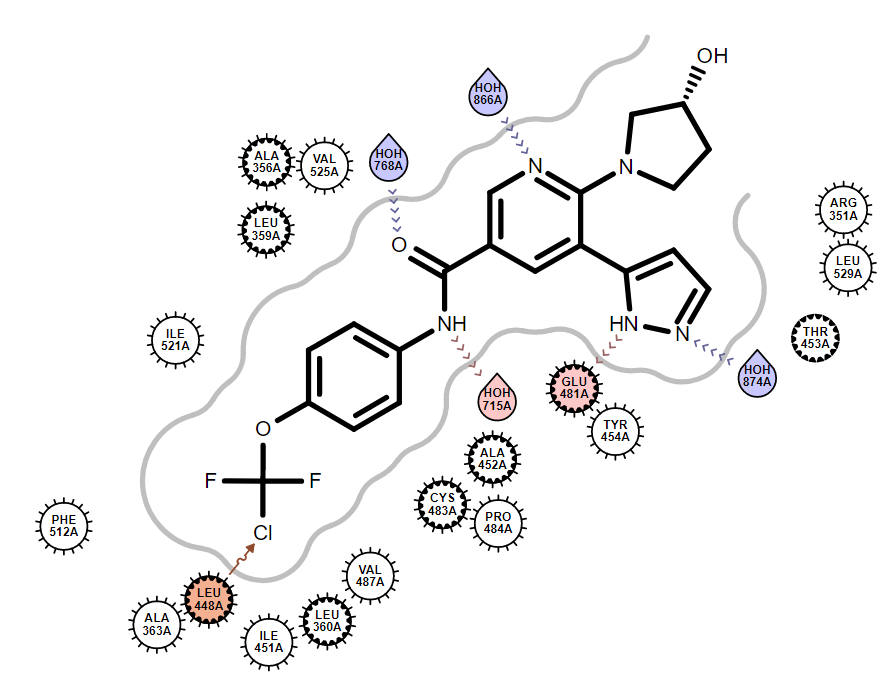 |
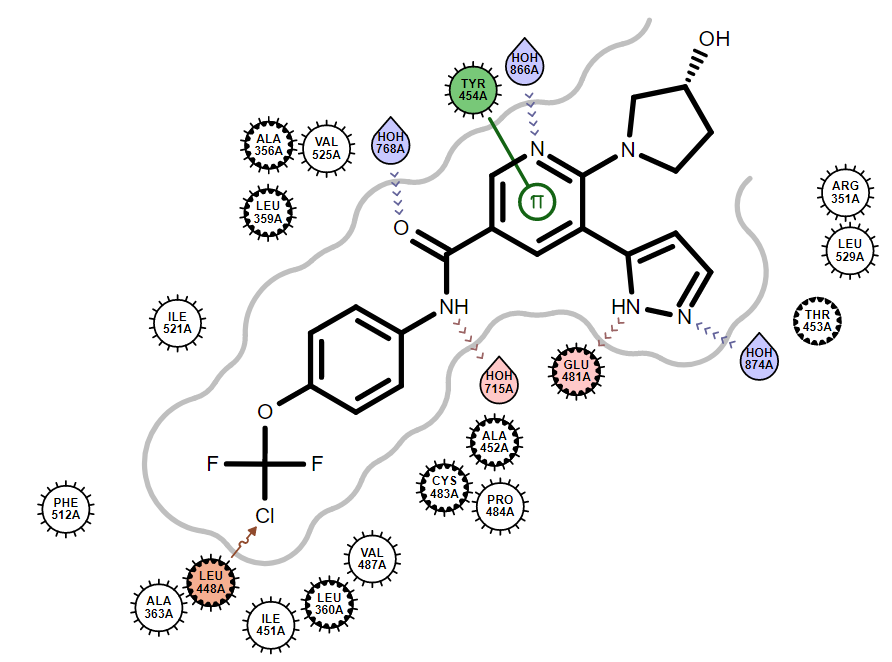 |
| 上节代码的执行结果 | 将面对边的pi-pi相互作用夹角放宽10度的执行结果 |
输出文字版的相互作用信息
大多数很多情况下,你只是需要将相互作用可视化,或者判断是否有存在预想的相互作用,而不需要输出所有的相互作用类型。如果您确实需要输出全部的信息,那也是很简单的。
首先先定义几个函数以便将相互作用信息打印出来:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 | def GetResidueName(residue): return "%3s %4d %s" % (residue.GetName(), residue.GetResidueNumber(), residue.GetChainID()) def GetInteractionString(inter): fragstrs = [] for frag in [inter.GetBgnFragment(), inter.GetEndFragment()]: if frag is None: continue fragtype = frag.GetComponentType() if fragtype == oechem.OELigandInteractionHintComponent(): fragstrs.append("ligand:" + " ".join(sorted(str(a) for a in frag.GetAtoms()))) if fragtype == oechem.OEProteinInteractionHintComponent(): fragstrs.append("protein:" + " ".join(sorted(str(a) for a in frag.GetAtoms()))) return " ".join(sorted(f for f in fragstrs)) def PrintResidueInteractions(asite, residue): ligatomnames = set() for inter in asite.GetInteractions(oechem.OEHasResidueInteractionHint(residue)): ligfrag = inter.GetFragment(oechem.OELigandInteractionHintComponent()) if ligfrag is None: continue for latom in ligfrag.GetAtoms(): ligatomnames.add(str(latom)) if len(ligatomnames) != 0: print(GetResidueName(residue), ": ", " ".join(sorted(a for a in ligatomnames))) def PrintLigandAtomInteractions(asite, atom): resnames = set() for inter in asite.GetInteractions(oechem.OEHasInteractionHint(atom)): profrag = inter.GetFragment(oechem.OEProteinInteractionHintComponent()) if profrag is None: continue for patom in profrag.GetAtoms(): residue = oechem.OEAtomGetResidue(patom) resnames.add(GetResidueName(residue)) if len(resnames) != 0: print(atom, ":", " ".join(sorted(r for r in resnames))) |
将分子内、分子间的各种相互作用类型(包括各种contact)全部打印出来:
1 2 3 4 5 6 7 8 9 | print("Number of interactions:", asite.NumInteractions()) for itype in oechem.OEGetActiveSiteInteractionHintTypes(): numinters = asite.NumInteractions(oechem.OEHasInteractionHintType(itype)) if numinters == 0: continue print("%d %s :" % (numinters, itype.GetName())) inters = [s for s in asite.GetInteractions(oechem.OEHasInteractionHintType(itype))] print("\n".join(sorted(GetInteractionString(s) for s in inters))) |
结果如下:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 | Number of interactions: 128 80 bio:active-site:contact : ligand: 0Cl protein:5796 C ligand: 0Cl protein:5797 O ligand: 0Cl protein:5799 C ligand: 0Cl protein:5800 C ligand: 0Cl protein:5856 C ligand: 0Cl protein:5858 C ligand: 0Cl protein:5871 N ligand: 0Cl protein:5875 C ligand: 0Cl protein:6428 C ligand: 3 C protein:4264 N ligand: 6 C protein:6345 C ligand: 6 C protein:6346 O ligand: 6 C protein:7101 C ligand: 7 N protein:7754 O ligand: 8 C protein:7031 C ligand: 8 C protein:7400 O ligand: 8 C protein:7754 O ligand: 9 C protein:5904 C ligand: 9 C protein:7031 C ligand: 9 C protein:7400 O ligand:10 C protein:5874 O ligand:10 C protein:5904 C ligand:10 C protein:6342 O ligand:10 C protein:7208 O ligand:11 C protein:6342 O ligand:12 C protein:5874 O ligand:12 C protein:6342 O ligand:13 C protein:4389 C ligand:13 C protein:7031 C ligand:13 C protein:7400 O ligand:14 N protein:7208 O ligand:15 O protein:4342 C ligand:15 O protein:4345 C ligand:15 O protein:4389 C ligand:15 O protein:7031 C ligand:15 O protein:7400 O ligand:16 C protein:7032 C ligand:16 C protein:7208 O ligand:17 C protein:4342 C ligand:17 C protein:4387 C ligand:17 C protein:4389 C ligand:18 C protein:4387 C ligand:18 C protein:6976 C ligand:19 C protein:6976 C ligand:20 C protein:5872 C ligand:20 C protein:5875 C ligand:20 C protein:6366 S ligand:20 C protein:6378 C ligand:20 C protein:6428 C ligand:21 C protein:5872 C ligand:21 C protein:5875 C ligand:21 C protein:6362 C ligand:21 C protein:6365 C ligand:21 C protein:6366 S ligand:21 C protein:6378 C ligand:21 C protein:7208 O ligand:22 O protein:6428 C ligand:22 O protein:6429 C ligand:22 O protein:6976 C ligand:24 N protein:5874 O ligand:24 N protein:6341 C ligand:24 N protein:6342 O ligand:25 N protein:5882 C ligand:25 N protein:7778 O ligand:26 C protein:5882 C ligand:26 C protein:5883 C ligand:26 C protein:5884 O ligand:27 C protein:4264 N ligand:27 C protein:5902 C ligand:28 C protein:6428 C ligand:29 F protein:4408 C ligand:29 F protein:4463 C ligand:29 F protein:6837 C ligand:30 F protein:4385 C ligand:30 F protein:4386 O ligand:30 F protein:4387 C ligand:30 F protein:4402 N ligand:30 F protein:4403 C ligand:30 F protein:4463 C ligand:30 F protein:5858 C 3 bio:active-site:hbond:protein2ligand : ligand: 7 N protein:7754 O ligand:15 O protein:7400 O ligand:25 N protein:7778 O 2 bio:active-site:hbond:ligand2protein : ligand:14 N protein:7208 O ligand:24 N protein:6342 O 25 bio:active-site:hbond:protein-intra-molecular : protein:4262 N protein:7625 O protein:4314 O protein:5906 O protein:4330 O protein:4383 N protein:4341 N protein:7883 O protein:4344 O protein:4402 N protein:4354 O protein:4421 N protein:4386 O protein:4459 N protein:4405 O protein:4469 N protein:4405 O protein:4474 O protein:5735 O protein:5794 N protein:5755 O protein:5813 N protein:5778 O protein:5852 N protein:5797 O protein:5871 N protein:5816 O protein:5886 O protein:5855 O protein:5895 N protein:5874 O protein:7208 O protein:5906 O protein:7400 O protein:6310 N protein:6364 O protein:6328 O protein:6361 N protein:6346 O protein:7592 O protein:6354 N protein:7658 O protein:6375 O protein:6423 N protein:6375 O protein:6439 N protein:7754 O protein:8135 O protein:8018 O protein:8135 O 13 bio:active-site:hbond:non-ideal-protein-intra-molecular : protein:4264 N protein:7871 O protein:4265 N protein:7625 O protein:4330 O protein:4367 N protein:4386 O protein:4442 N protein:5840 O protein:5881 N protein:5886 O protein:5923 N protein:6339 N protein:7778 O protein:6366 S protein:6375 O protein:6993 O protein:7883 O protein:7592 O protein:7871 O protein:7592 O protein:8210 O protein:7871 O protein:8210 O protein:7883 O protein:8135 O 1 bio:active-site:hbond:unpaired-ligand-acceptor : ligand:23 O ligand:23 O 2 bio:active-site:hbond:unpaired-protein-acceptor : protein:5884 O protein:5884 O protein:6357 O protein:6357 O 1 bio:active-site:halogen:protein-nucleophile : ligand: 0Cl protein:5797 O 1 bio:active-site:halogen:ligand-intra-molecular : ligand: 0Cl ligand:22 O |
可以发现,总共发生了128个相互作用,并列出了具体的相互作用类型,比如其中第136行就显示出在蛋白与配体之间有一个卤键相互作用。这个相互作用我们可以从生成的图片里找到。除了分子内相互作用之外,其它的OEChem计算的相互作用已经呈现在Oedepict绘制的图片里面了。
拓展应用
OEChem强大的相互作用分析不仅可以用于视觉分析,还可以用来对docking结果进行后处理,比如过滤出满足特定相互作用模式的化合物。




