摘要:Flare V6的一个新特性是引入了配体QM计算。本文以(S)-Dexmedetomidine为例介绍了如何通过Flare调用PSI4进行ECD光谱计算,具体步骤包括:XED力场构象优化、XED力场构象搜索、QM构象优化、构象去重、激发态计算以及ECD图谱绘制。
肖高铿/2022-09-22
简介
Flare V6是一款集成了基于结构、基于配体与QM计算的综合性软件包,并引入多个新的特性[1,2,3],其中一个新特性是配体分子的QM计算。之前已经介绍过,如何在Flare里用QM研究配体的静电特性[1]与构象稳定性[4]。本文以(S)-Dexmedetomidine(图1化合物)为例介绍如何通过Flare的Jupyter界面里调用PSI4[5]进行ECD光谱计算,详细的PSI4计算ECD图谱还可参见PSICON 2020的TDDFT教程[6]。本文的目的是演示,而非科研,因此更详细的ECD光谱计算规范请参考Pescitelli等人的文献[7]。
图1. (S)-Dexmedetomidine
Dexmedetomidine的SMILES代码如下:
1 | C[C@@H](c1cccc(C)c1C)C2=CNC=N2 |
XED力场优化与构象搜索
首先,我们用Flare | Ligand | New创建(S)-Dexmedetomidine的3D结构(图2 步骤1),并视觉检查确保立体化学信息是正确的。然后用Flare | 3D Pose | Minimize使用XED对创建的结构进行精确优化(图2 步骤2与3)。
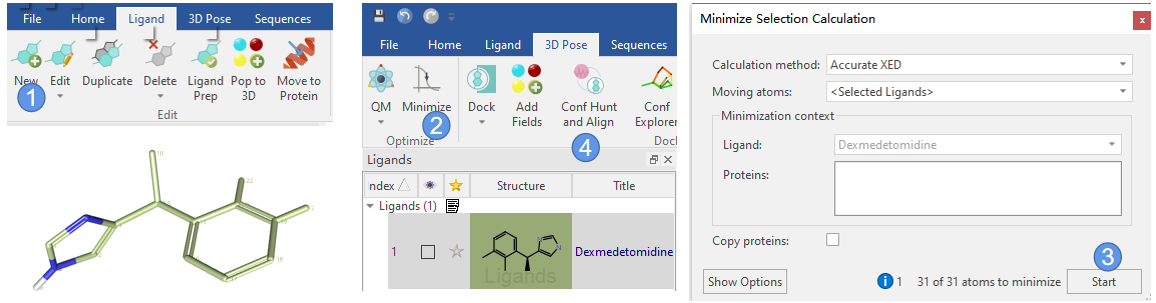
图2. 创建3D结构与XED力场优化
有了XED力场优化过后的3D构象,我们开始用XED力场进行构象搜索。点击 Flare | 3D Pose | Conf Hunt & Align (图2 步骤4),弹出图3所示的构象搜索与叠合对话框,使用如下参数:
- Conformation Hunt: Very accurate and slow
- Alignment:No Calculation
- Ligands to Conf hunt & align:Selected ligand
然后点击start开始构象搜索,见图3步骤5。
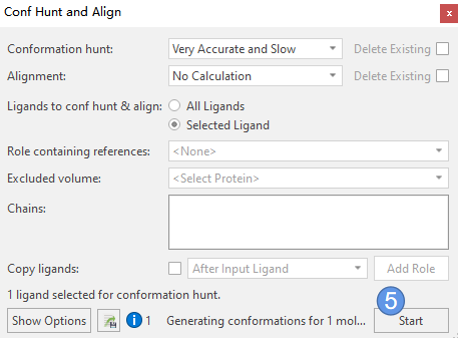
图3. Conf Hunt & Align对话框
构象搜索结束,用Flare | 3D Pose | Conf Explorer可以发现总共找到了12个低能构象(能量窗=3kcal/mol,见图4左),这12个构象的侧视图与顶视图如图4所示。
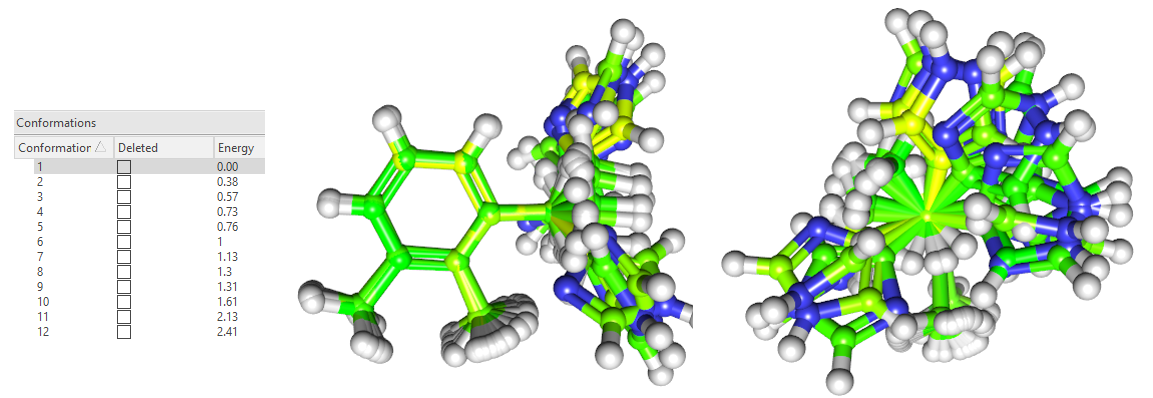
图4. 12个低能构象
用QM优化构象
接下来用QM进一步对每个构象进行结构优化。点击Flare | 3D Pose| QM | Minimize Conformations(图5步骤6),弹出QM Minimize Conformers对话框,设置参数如下(图5步骤7)。优化分两个步骤进行:优化与单点能计算。
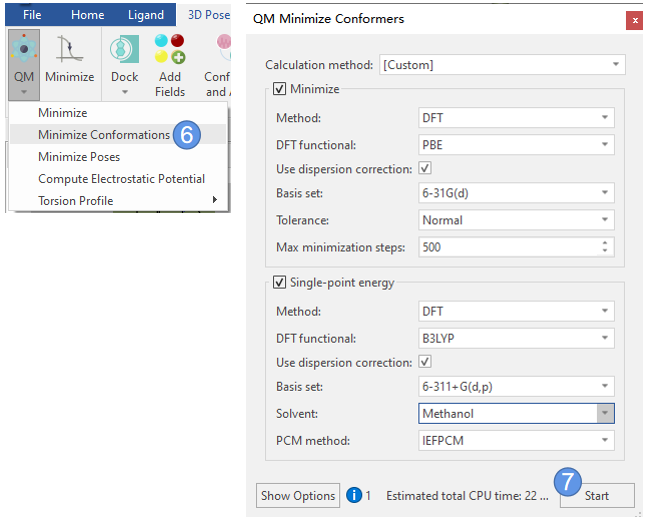
图5. QM优化构象
其中优化步骤参数如下:
- Method: DFT
- DFT functional:PBE
- Use dispersion correction:Yes
- Basis Set:6-31G(d)
- Tolerance:normal
- Max minimization steps:500
其中单点能计算步骤参数如下:
- Method: DFT
- DFT functional:B3LYP
- Use dispersion correction:Yes
- Basis Set:6-311+G(d,p)
- Solvent:Methanol
- PCM Model:IEFPCM
其中单点能计算的能量将用于构象系综分析,计算玻尔兹曼权重。溶剂的选择与实验数据采用的溶剂一致,也与下一步激发计算ECD图谱的溶剂一致。
计算完毕,点击Flare | 3D Pose | Conf explorer(图6步骤8)可以浏览每个优化的构象以及对应的XED与QM构象能。
图6. 构象分析Conf explorer对话框
在用QM优化之后,如图7所示,我们发现QM能量排序与XED力场能量排序不同,而且还有几个化合物的QM能量都为0。这提示,可能有部分构象优化之后得到相同的构象。为了减少后续TDDFT激发态计算的计算量,有必要将相同的构象合并(去重复)。
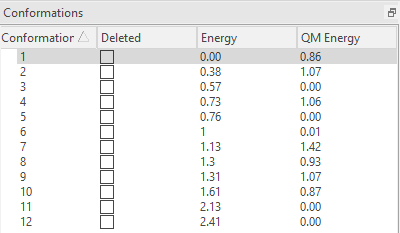
图7. QM构象能
在这个例子中,构象5与3重复,构象12与11重复,因此选择5与12(图8步骤8),先点击Del(图8步骤9),再点击accept则删除构象5与12(图8步骤10)。
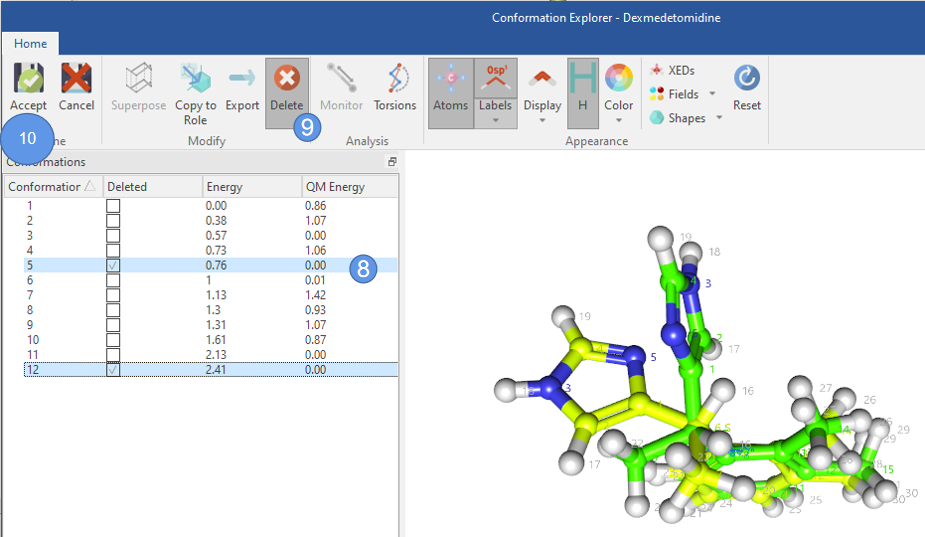
图8. 构象去重复
最后,我们得到7个不重复的构象,将这些优化好的构象导出,保存为sdf文件备用。这个算例里,我将7个独立不重复的构象分别保存单个的SDF文件,共7个文件,分别为:CONF_1.sdf, CONF_2.sdf…,CONF_7.sdf。
激发态计算:ECD图谱生成
新建一个Flare project,点击Python | Jupyter打开Jupyter notebook。现在以最低能的构象CONF_1.sdf为例说明ECD光谱的计算,其它构象的ECD光谱计算采用完全一样的方法。
首先切换工作目录到保存构象的目录:
1 2 3 | from cresset import flare import os os.chdir('/public/gkxiao/work/flare_ecd') |
从SDF文件里提取分子的坐标,以便构造ECD计算的分子描述:
1 2 3 4 5 6 7 8 9 10 11 | from rdkit import Chem mol = Chem.MolFromMolFile('CONF_01.sdf',removeHs=False) n = mol.GetNumAtoms() formalcharge = str(Chem.rdmolops.GetFormalCharge(mol)) for i in range(n): pos = mol.GetConformer().GetAtomPosition(i) element = mol.GetAtoms()[i].GetSymbol() x = str(round(pos.x,4)) y = str(round(pos.y,4)) z = str(round(pos.z,4)) print(element+" "+x+" "+y+" "+z) |
输出如下:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 | C -0.9189 3.0772 0.5403 C -1.7467 4.0246 1.1222 N -1.5394 5.1889 0.4001 C -0.6108 4.9054 -0.5698 N -0.2164 3.6429 -0.5151 C -0.7283 1.6284 0.9049 C -1.2646 1.3199 2.317 C -1.3523 0.665 -0.1126 C -2.4974 1.0673 -0.8235 C -3.1342 0.2034 -1.7173 C -2.6248 -1.0845 -1.9131 C -1.4857 -1.5177 -1.2171 C -0.8422 -0.6433 -0.3022 C 0.3692 -1.1625 0.441 C -0.9554 -2.9135 -1.445 H 0.3649 1.4676 0.9034 H -2.4391 3.9861 1.9596 H -1.9863 6.0863 0.5731 H -0.2616 5.6557 -1.2793 H -1.0578 0.273 2.5925 H -2.3595 1.4573 2.352 H -0.8032 1.9801 3.0717 H -2.8794 2.0821 -0.6748 H -4.0222 0.5351 -2.2656 H -3.1126 -1.7694 -2.6162 H 1.1553 -1.498 -0.2602 H 0.8218 -0.4154 1.1075 H 0.1109 -2.0414 1.0609 H 0.0834 -2.903 -1.8244 H -0.9401 -3.5072 -0.512 H -1.5738 -3.4564 -2.1779 |
接着在B3LYP/6-311+G(d,p)理论水平下进行激发态计算,并以IEFPCM为溶剂模型考虑甲醇的溶剂效应:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 | import psi4 import numpy as np from psi4.driver.procrouting.response.scf_response import tdscf_excitations # Output file path # arg 0: Path of output file # arg 1: Whether overwrite or not psi4.core.set_output_file("CONF_1_tddft.out",True) # set threads and memory psi4.core.set_num_threads(24) psi4.set_memory(int(24e9)) # Scratch directory path psi4.core.IOManager.shared_object().set_default_path("./tmp") pcm_string = """ Units = Angstrom Medium { SolverType = IEFPCM Solvent = Methanol Nonequilibrium = True } Cavity { Type = GePol Area = 1.0 } """ # molecule CONF_1 = psi4.geometry("""0 1 C -0.91890 3.07720 0.54030 C -1.74670 4.02460 1.12220 N -1.53940 5.18890 0.40010 C -0.61080 4.90540 -0.56980 N -0.21640 3.64290 -0.51510 C -0.72830 1.62840 0.90490 C -1.26460 1.31990 2.31700 C -1.35230 0.66500 -0.11260 C -2.49740 1.06730 -0.82350 C -3.13420 0.20340 -1.71730 C -2.62480 -1.08450 -1.91310 C -1.48570 -1.51770 -1.21710 C -0.84220 -0.64330 -0.30220 C 0.36920 -1.16250 0.44100 C -0.95540 -2.91350 -1.44500 H 0.36490 1.46760 0.90340 H -2.43910 3.98610 1.95960 H -1.98630 6.08630 0.57310 H -0.26160 5.65570 -1.27930 H -1.05780 0.27300 2.59250 H -2.35950 1.45730 2.35200 H -0.80320 1.98010 3.07170 H -2.87940 2.08210 -0.67480 H -4.02220 0.53510 -2.26560 H -3.11260 -1.76940 -2.61620 H 1.15530 -1.49800 -0.26020 H 0.82180 -0.41540 1.10750 H 0.11090 -2.04140 1.06090 H 0.08340 -2.90300 -1.82440 H -0.94010 -3.50720 -0.51200 H -1.57380 -3.45640 -2.17790 """, name="CONF_1") psi4.set_options({ "save_jk": True, "pcm": True, "pcm__input": pcm_string }) e, wfn = psi4.energy("B3LYP-D3/6-311+g(d,p)", return_wfn=True, molecule=CONF_1) res = tdscf_excitations(wfn, states=100) |
计算完毕,可以将波长(激发能)、振子强度、转子强度打印出来以便用绘图工具绘制UV与ECD图谱。
1 2 3 4 5 6 7 8 9 | import numpy as np # get poles and residues to plot OPA and ECD spectra poles = [r["EXCITATION ENERGY"] for r in res] opa_residues = [np.linalg.norm(r["ELECTRIC DIPOLE TRANSITION MOMENT (LEN)"])**2 for r in res] ecd_residues = [r["ROTATORY STRENGTH (LEN)"] for r in res] print(poles) print(opa_residues) print(ecd_residues) |
将poles(激发能量,也就是波长), opa_residues(振子强度)与ecd_residues(转子强度)三个数组打印,结果如下:
1 2 3 | [0.1863171096523767, 0.1939039458434858, 0.19659912134475524, 0.2050404252248169, 0.2078159462592276, 0.21483312155800743, 0.21558247810441128, 0.21629287700878633, 0.22131073136342963, 0.2239578317414286, 0.2256655366089712, 0.2266063071494987, 0.22719511927490812, 0.22912871681783578, 0.23036097855538518, 0.23196639438645655, 0.23288762018732767, 0.23501998580463102, 0.23608956077502005, 0.23782839085338345, 0.24087960247833654, 0.24129619231009988, 0.24312235507952845, 0.24410980694671544, 0.24589867812187582, 0.24602973193576907, 0.2474711509867622, 0.24806441736255788, 0.24915399324363377, 0.2505334198822149, 0.2507468669141498, 0.2516462192853461, 0.2529149024536076, 0.2539699801159058, 0.25493797633166865, 0.25604830696224573, 0.256712318336855, 0.25910518815857936, 0.25934435981982784, 0.26037322293828696, 0.26064815699912447, 0.26262871872268084, 0.26279939036390276, 0.26427965834506684, 0.2648107191555363, 0.2651422483600908, 0.2669828436118275, 0.26821836857864967, 0.26923280469950706, 0.2700501638430005, 0.2716009041123941, 0.2726448138481998, 0.2732880554160093, 0.2736440419875829, 0.274309933278269, 0.2744231250304597, 0.2746492115721138, 0.2754770447976371, 0.27620293298686066, 0.2769771531293081, 0.27768323690960944, 0.2785993363606018, 0.2812450116133058, 0.28204969870853414, 0.2829438615368195, 0.2829959857005867, 0.28372193197946083, 0.2850708970415412, 0.28577833597122376, 0.28654236330135197, 0.2876612810257548, 0.28882846369903425, 0.2889687498366261, 0.29223770672702615, 0.2924971774299208, 0.29301078633501404, 0.2935533342680961, 0.29517983860033936, 0.2959742796563639, 0.29693311006244916, 0.2978151847572293, 0.2989882344717956, 0.29912882378353234, 0.3003095203736919, 0.3004241483808328, 0.3006209011700486, 0.3014339300694061, 0.3020126831499768, 0.3028025674303158, 0.30344091532026335, 0.3038045324249683, 0.30432506025932426, 0.30542062626395344, 0.305848061297033, 0.3065622201511003, 0.3073873287851144, 0.30803302494442214, 0.30875256479223057, 0.3091063752678208, 0.3093240808604151] [0.008660083691178442, 0.10349305939435005, 0.048931279920771016, 0.01664861869646239, 0.023607697352347273, 0.4331760971827799, 0.19722087693559762, 0.1610869707851778, 0.5004291874968285, 0.5362120773474215, 0.7385523305745509, 1.0827967793656208, 1.0953285186803543, 1.2331583251069618, 1.7809304053128256, 0.9159936256376622, 0.15939112329899552, 1.2124717270895917, 0.08368402618706905, 0.6349603125759534, 0.16955022615511947, 0.004088688844779394, 0.11457343408462559, 0.5537920767918361, 0.008431317491542988, 0.055311354295249686, 0.09490812203280845, 0.14436463677335507, 0.12293991809471337, 0.03846757998370501, 0.04355763322776323, 0.03549218640806217, 0.018829640362198714, 0.015675566949828388, 0.06707526979885038, 0.0472314396469641, 0.001410585141789898, 0.027249991469494337, 0.01215669411084908, 0.02896998022629227, 0.023703964733904008, 0.05161528208433824, 0.06505625050737211, 0.06650963753683756, 0.3069988672801764, 0.024996907357562683, 0.013209539481416309, 0.052133675150076955, 0.049847755758164566, 0.26341816888027014, 0.007457044778791769, 0.005532852603604105, 0.0029111373742524587, 0.13346151626882374, 0.07063898473568661, 0.05371989166671687, 0.020279016086313446, 0.05534948465653796, 0.13935646199510285, 0.10236858715359076, 0.03602978482437635, 0.12279445737442111, 0.0011384301920499532, 0.1669445184936711, 0.13900267330987612, 0.003922858485403657, 0.12277084337674618, 0.0901787539114518, 0.08064482953366019, 0.0012033748240366372, 0.05832423778338989, 0.026998761335166, 0.3057183938678913, 0.15798173989573985, 0.055852891878045915, 0.17497903314275592, 0.037460505497793214, 0.09543486080372354, 0.023497561837022337, 0.04359603931558043, 0.07871790650307069, 0.050023076497197116, 0.04721549532703174, 0.001442058515559898, 0.007704308350750135, 0.032359280805689984, 0.030062327596333697, 0.1423759674141197, 0.30719405791743637, 0.08069097181483685, 0.10063493272630698, 0.09834923258949534, 0.02403182544587331, 0.18789039951589168, 0.021025667764258474, 0.03665697332342831, 0.11860141677048568, 0.013461398062255187, 0.023357460709835222, 0.008771811915421562] [-0.0023185573599461603, 0.007772880795271462, -0.023006054965657698, 0.02823403191495033, -0.0030940906925422634, 0.033226070133156656, 0.026959457729096383, -0.007603433402276863, -0.0033942796205106507, 0.12709248350914848, 0.006601752756492565, -0.12323723569666853, 0.09016329672615413, -0.17172020363322477, 0.09223222988505309, 0.11378531083618075, -0.018570489116571665, 0.07928101788937807, -0.0362165571670183, -0.17683827015687828, -0.03409388329565155, 0.002638020701523085, 0.030585166885200642, 0.021051040289371256, 0.008822610641127486, -0.012552096713053363, 0.03546296455003074, -0.027230305756463534, -0.04345447982360856, 0.018979307688783805, -0.0205165787254387, 0.005159670113488453, -0.002044918212805822, 0.00013773234326517456, -0.028981342251422307, -0.014244631324884715, -0.001852254650470736, -0.016698127679452134, 0.010445765682826128, 5.988789893958402e-05, 0.0055470568610092585, 0.01099414941510461, -0.024311972717794596, -0.025265724796203617, 0.0037465736174804986, -0.002780122636042534, 0.008249586231930037, -0.018901303345790908, 0.013273229076012794, 0.0034774907690217015, -0.012259300424381028, -0.00038453055996658765, 0.0013796841993837599, 0.02460696753483636, 0.017809394239199774, 0.02147496433000713, 0.014033966718485457, 0.008078207634736823, 0.024635290576418802, 0.04407696637364731, -0.009629366290395465, -0.111195034016115, 0.0010305382280823652, -0.03962034048826819, 0.022109867011012085, -0.003894319272733941, -0.009897340425816765, 0.01983557425706388, 0.026573444857911882, 0.001187866266289116, 0.01631066940269573, 0.005504560000458998, -0.03100658728361818, -0.008949408647276294, 0.05188512541525664, -0.013211560802535487, -0.02509309024064607, -0.013474468990270466, 0.0058157801608803225, 0.00941595475192742, -0.001600915522050994, 0.0006496899062446836, 0.00570793877230435, -0.0015902489599031139, 0.0020201232011859624, 0.02135661646865954, -0.02734191973113685, 0.0558643389082213, -0.025157937409131477, 0.03787978186567708, -0.02756321082487943, -0.009788330784436538, 0.01812071052000217, 0.008672496271963774, -0.00023955892922729403, 0.0008040016414064197, -0.01293985186918948, 0.006059973401203619, 0.034157745289823654, -0.0025525230946705493] |
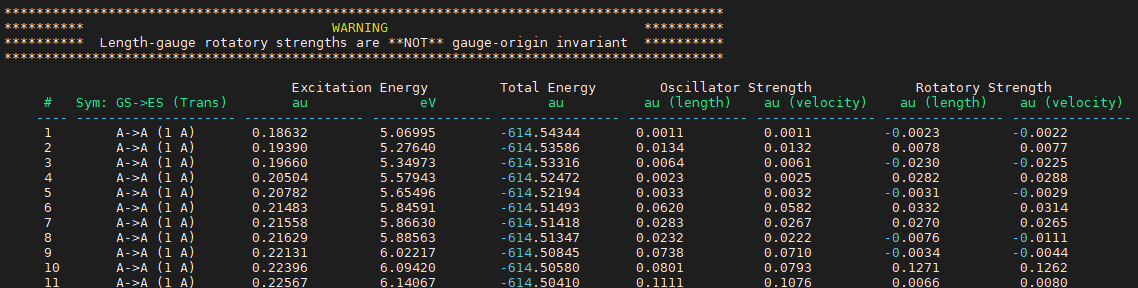
poles, opa_residues与ecd_residues的信息还可以在输出文件CONF_1_tddft.out找到,部分结果如下图9所示。
图9. 激发态计算结果文件里关于激发能、振子强度与转子强度的信息,这里只是呈现了前10个跃迁。
有了激发能、振子强度与转子强度,就可以绘制UV与ECD图谱,代码如下:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | import numpy as np from rdkit import Chem from rdkit.Chem import altair_spectrum from altair_spectrum import plot_spectrum from psi4.driver.p4util import spectrum opa_spectrum = spectrum(poles=poles, residues=opa_residues, gamma=0.01, out_units="nm") ecd_spectrum = spectrum(poles=poles, residues=ecd_residues, kind="ECD", gamma=0.01, out_units="nm") opa_plot = plot_spectrum(opa_spectrum, title="OPA (Gaussian broadening)", x_title=("λ", "nm")) ecd_plot = plot_spectrum(ecd_spectrum, title="ECD (Gaussian broadening)", x_title=("λ", "nm"), y_title=("Δε", "L⋅mol⁻¹⋅cm⁻¹")) opa_plot & ecd_plot |
激发态计算相当快,在我的24核心机器上大约1小时不到就完成了。计算图谱如图10所示,需要注意的是:sigma值是经验值,这意味着需要调整该值以使得计算图谱与实验图谱尽可能一致。
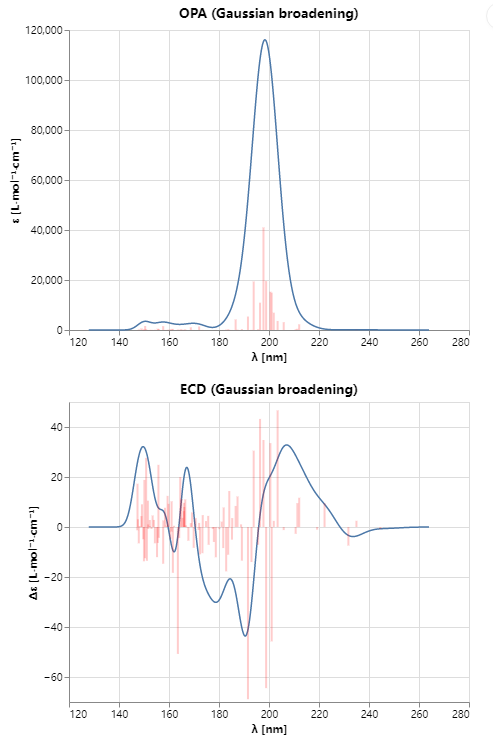
图10. 构象1的计算ECD图谱。上:UV谱;下:ECD谱。
每个构象都有一个自己的ECD图谱,需要进一步采用玻尔兹曼加权平均法获得多个构象的平均图谱,最后将实验图谱与平均图谱进行比较以确认立体化合物构型。具体过程请参见前文[8],这里不再叙述。
小结
总的来说,Flare集成了ECD计算所需的全部工具,可以一步到位的实现3D结构创建、XED力场结构优化、XED力场构象搜索、QM构象优化、激发态计算与UV、ECD图谱绘制。整个过程操作便利、容易实现、而且计算高效。Flare是非常实用的ECD计算辅助结构确证平台。
文献
- Flare V6发布——新特性、新功能与改进. http://blog.molcalx.com.cn/2022/07/06/flare-v6-released.html
- Flare V6预告——计算化学先睹为快. http://blog.molcalx.com.cn/2022/05/03/sneak-peek-flare-v6-comp-chem.html
- Flare V6预告——药化先睹为快. http://blog.molcalx.com.cn/2022/04/26/sneak-peek-flare-v6-medicinal-chemists.html
- 势能面扫描在Torsion Profile中的应用. http://blog.molcalx.com.cn/2020/01/30/qm-torsion-profile.html
- PsiCode-PSI4: Ab Initio Quantum Chemistry. https://psicode.org
- Roberto Di Remigio. TDDFT tutorial at PsiCon2020. https://github.com/robertodr/tddft_psicon2020
- Pescitelli, G.; Bruhn, T. Good Computational Practice in the Assignment of Absolute Configurations by TDDFT Calculations of ECD Spectra. Chirality 2016, 28 (6), 466–474. https://doi.org/10.1002/chir.22600.
- ECD图谱预测与生成Excel报告. http://blog.molcalx.com.cn/2020/01/01/ecd.html