摘要:Lead Finder是一款分子对接软件,本文以TK激酶抑制剂的虚拟筛选为例,演示了如何用Lead Finder对化合物数据库进行基于分子对接的虚拟筛选流程:1)蛋白结构的下载、预处理;2)用build_model来进行蛋白结构的准备;3)用Lead Finder进行能量格点的准备;4)Lead Finder进行分子对接计算、虚拟筛选与并行计算;4)虚拟筛选性能评价等。

一. Lead Finder简介
1. Lead Finder的用途
Lead Finder™是一款新颖的分子对接软件,为满足计算化学与药物化学科研人员在药物发现过程的需求而设计。此外,它不仅适用于涉及ADMET的药理、毒理科研人员,还适用于研究酶的专一性、理性酶设计的生物与酶学专家。
用Lead Finder您可以:
- 通过虚拟筛选发现新颖的先导化合物
- 通过虚拟筛选设计高度富集活性化合物的集中库
- 将配体共价或非共价对接到蛋白上
- 快速评估新分子与蛋白活性位点的匹配性
2. Lead Finder分子对接精度
跟据测试,Lead Finder比同行GOLD,GLIDE,FlexX, Surflex-Dock在同样的数据集上显著地表现出更加优秀的结合模式预测性能(re-docking精度)。
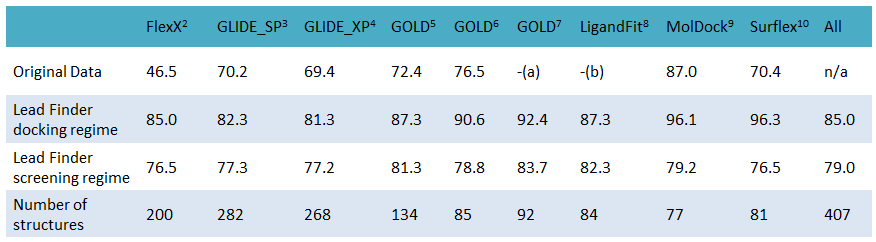
上表数据出处:BioMolTech,Original Data行为对应列软件的对接精度;Lead Finder行的每列对应着Lead Finder用相应软件测试集测试获得的性能结果。
3. Lead Finder分子对接的虚拟筛选性能
二. 虚拟筛选流程
1. 算例介绍
胸苷激酶(Thymidine kinase)是一种磷酸转移酶(激酶), 存在于大部分活体细胞中。它以两种同工酶的形式存在于哺乳动物细胞中,TK1和TK2。TK是HSV感染首选药阿昔洛韦的治疗靶。阿昔洛韦可被病毒胸腺嘧啶激酶(Thymidine Kinase, TK)催化磷酸化变成一磷酸无环鸟苷(acyclo-GMP),此过程比细胞胸腺嘧啶激酶快3000多倍。在细胞酶的催化下转变成二磷酸无环鸟苷(acyclo-GDP)及三磷酸无环鸟苷(acyclo-GTP)。acyclo-GMP是一种十分有效的DNA聚合酶抑制剂,它与病毒的亲和力是细胞聚合酶的100倍。作为酶作用物,acyclo-GTP与病毒的DNA结合,导致DNA链终止。同时,病毒酶不能将acyclo-GTP从DNA链上除去,从而导致DNA聚合酶的作用被进一步的抑制。(引用自Wiki:阿昔洛韦)
本文以TK为例,用10个已知的Tk抑制剂以及几百个Decoy化合物来演示虚拟筛选的操作步骤以及虚拟筛选性能评估等。
2. Lead Finder的软件组成
Lead Finder软件包包含build_model与leadfinder两个子程序:前者负责蛋白结构准备;后者负责计算能量格点与分子对接。
Build Model是Lead Finder里准备蛋白的计算模块,它根据Mehler E. L.的SCP理论1,2在指定的pH值下归属蛋白残基离子化状态、准备蛋白结构,包括:(1)根据给定的pH值优化残基的离子化状态对蛋白加氢;(2)根据出现在蛋白里的配体、辅酶、底物优化极性氢的取向;(3)优化His、Asn与GLn残基的侧链取向;(4)残基的侧链重建;(5)氨基酸残基突变;(6)单个残基GAP的填充。
3. 操作步骤
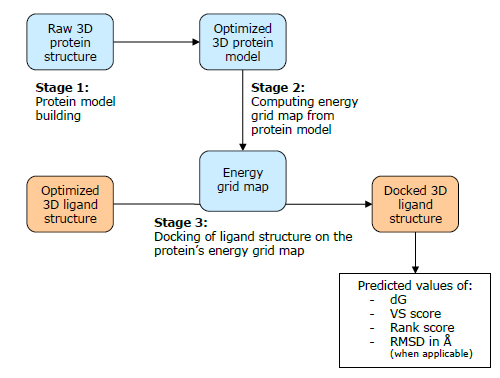
图1. Lead Finder分子对接操作流程
步骤1. 蛋白结构的准备
从PDB数据库上下载PDB code为1KIM的蛋白晶体结构1kim.pdb。该晶体结构为二聚体,含两条一模一样的亚基链A、B,每条链各含一个配体THM。我们只需要用A链即可,因此要删除1kim.pdb里的B链、A链的硫酸根离子,并保存为1kim_ChainA.pdb:
1 | grep '.....................A' 1kim.pdb |grep -v SO4 >> 1kim_ChainA.pdb |
准备参数文件1kim.set,内容如下:
1 | ligand res name THM |
现在开始用Build Model准备蛋白结构:
1 | build_model -f 1kim_ChainA.pdb -set 1kim.set -olog model.log |
生成配体(ligand_reference.pdb)与蛋白文件(protein.pdb)
1 | 1kim_ChainA.pdb 1kim.pdb 1kim.set ligand.mol ligand_reference.pdb model.log protein.pdb |
步骤2. 生成能量格点
1 | leadfinder --grid-only --protein=protein.pdb --ligand-reference=ligand_reference.pdb --save-grid=protein.grid |
步骤3. 虚拟筛选(分子对接)
准备工作:
tk.sdf: 10个已知的TK抑制剂,源自Surflex-Dock测试文件;
decoy.sdf: 10个已知的TK抑制剂的decoy化合物,源自Surflex-Dock测试文件。
分子对接能量格点文件:protein.grid
TK抑制剂的分子对接:
1 | leadfinder -g protein.grid -li tk.sdf -os ligand_score.csv -l ligand_report.log -mp 20 -rta -rtc all -rte |
TK抑制剂decoy的分子对接:
1 | leadfinder -g protein.grid -li decoy.sdf -os decoy_score.csv -l decoy_report.log -mp 20 -rta -rtc all -rte |
关于Lead Finder的并行计算
- 指定核心数进行计算:-np N, –number-of-processes=N
- 给计算节点分配指定的化合物数量:-js N, –mpi-job-size=N
N:指定参与计算的核心数。该选项适于单机多核或集群环境计算。注意:1)需要license支持;2)仅在命令行模式下可用
N: 集群环境下,在管理节点从计算节点回收计算结果前计算节点应该完成的化合物处理量,管理节点根据其自己的作业列表每次从计算节点归拢计算结果。默认值为100。注意:仅适用于命令行模式。
比如用48个核心在虚拟筛选模式(vs)下并行计算:
1 | leadfinder -vs -np 48 -g protein.grid -li decoy.sdf -os decoy_score.csv -l decoy_report.log -rta -rtc all -rte |
比如用48个核心在虚拟精确模式(xp)下并行计算:
1 | leadfinder -xp -np 48 -g protein.grid -li decoy.sdf -os decoy_score.csv -l decoy_report.log -rta -rtc all -rte |
步骤4. 虚拟筛选的性能评估
每个分子都会被用三种打分打分函数打分:dG score, VS score与Rank score。dG score用于预测结合亲合力;VS score用于虚拟筛选对化合物排序用;Rank score用于结合模式预测排序用。我们要考察Lead Finder的虚拟筛选性能,因此采用VS score来绘制ROC曲线、计算ROC AUC与富集因子。具体的ROC曲线绘制、性能评估操作流程见教程:如何进行虚拟筛选的方法学验证
(1)ROC曲线

图2. Lead Finder虚拟筛选的ROC曲线
从图2可以看出,ROC曲线靠近左上角,具有显著的富集能力;同时,曲线的一段与做坐标重合,说明Lead Finder在此计算中显示出非常优秀的早期富集能力。
(2)ROC AUC与富集因子(Enrichment factor)
表1. ROC AUC与EF
| TOP X% | 0.5% | 1% | 2% | 5% | 10% | 100% |
|---|---|---|---|---|---|---|
| pROC AUCX% | 0.144 | 0.219 | 0.309 | 0.480 | 0.699 | 0.977 |
| EFX% | 40 | 40 | 20 | 16 | 10 | 1 |
其中EFMAX = 71.8
(3)命中率(Hit Rate)
为了方便比较,考察Top 5%的命中率:
Hit Rate5% = EF5% / EFMAX = 16/71.8 = 22.3%
根据Kellenberger等人在2004年测试,8种不同软件命中率(Top 5%)如下表。虽然各种软件已经有新版本出来、该数据不能代表现各个软件现在的性能,但足以说明Lead Finder具有很好好的虚拟筛选命中率。
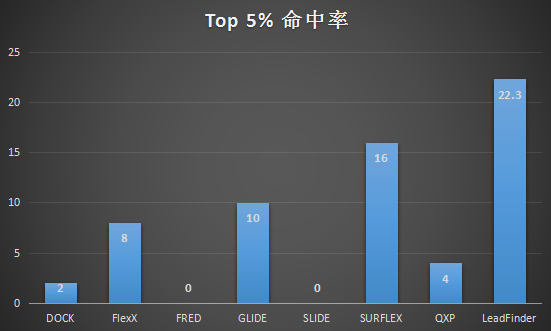
图3. TK激酶抑制剂虚拟筛选数据集Top 5%命中率
数据来自:Kellenberger, E.; Rodrigo, J.; Muller, P.; Rognan, D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 2004,57:225-42.
关于Lead Finder的打分输出文件
Lead Finder的os选项控制打分值输出文件的格式, 语法如下:
1 2 3 | -os FILENAME.[csv|txt] --output-tabular=FILENAME.[csv|txt] Output predicted values in tabular format (default FILENAME is energy.log) |
以csv格式输出的文件样式如下:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 | "Lead Finder version 1703 build 1, 25 March 2017" Docking summary Ligand,Rank,"dG, kcal/mol",VS score,"RMS, A",Docking time,Ligand Efficiency (dG/Nheavy),Molar weight,N(heavy atoms),N(FRBs),E(LJ),E(metal),E(sol),E(H-bonds),E(elect),E(internal),E(tors),E(dihedral),E(penalty),mol header,mol comment tk3d.sdf-ligand-1,0,-6.47514,-7.65557,N.A.,6.42,-0.404696,225.205,16,4,-2.51024,0,-4.17282,-0.344149,-0.213501,0.295181,0.4704,0,0,TK_aciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-1,1,-6.99137,-7.82896,N.A.,6.42,-0.436961,225.205,16,4,-2.35101,0,-4.23468,-1.08844,-0.10014,0.3125,0.4704,0,0,TK_aciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-1,2,-6.48715,-7.62512,N.A.,6.42,-0.405447,225.205,16,4,-2.47648,0,-4.20238,-0.394015,-0.235404,0.350733,0.4704,0,0,TK_aciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-1,3,-6.48741,-7.55464,N.A.,6.42,-0.405463,225.205,16,4,-2.46478,0,-4.16046,-0.433819,-0.225447,0.326692,0.4704,0,0,TK_aciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-2,0,-8.62618,-9.57187,N.A.,5.88,-0.479232,368.125,18,2,-4.16289,0,-5.05485,0.580639,-0.432822,0.208544,0.2352,0,0,TK_ahiu,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-2,1,-5.74903,-10.4789,N.A.,5.88,-0.31939,368.125,18,2,-4.10938,0,-4.97266,0.104522,-0.186232,3.17952,0.2352,0,0,TK_ahiu,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-2,2,-1.74842,-5.12982,N.A.,5.88,-0.0971346,368.125,18,2,-2.71733,0,-5.12118,0.29828,-0.0263348,5.58294,0.2352,0,0,TK_ahiu,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-2,3,-3.82981,-5.10346,N.A.,5.88,-0.212767,368.125,18,2,-0.493873,0,-5.14776,0.8253,0.0490504,0.702267,0.2352,0,0,TK_ahiu,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-3,0,-7.00542,-7.84963,N.A.,7.03,-0.467028,214.218,15,4,-2.28831,0,-4.42307,-0.634427,-0.376197,0.246181,0.4704,0,0,TK_dhbt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-3,1,-7.07601,-7.42948,N.A.,7.03,-0.471734,214.218,15,4,-2.37295,0,-4.33625,-0.796266,-0.304676,0.26374,0.4704,0,0,TK_dhbt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-3,2,-7.08851,-7.48931,N.A.,7.03,-0.472567,214.218,15,4,-2.40956,0,-4.38885,-0.772009,-0.216962,0.228475,0.4704,0,0,TK_dhbt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-3,3,-6.61773,-7.48222,N.A.,7.03,-0.441182,214.218,15,4,-2.26944,0,-4.34685,-0.44575,-0.244551,0.218459,0.4704,0,0,TK_dhbt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-3,4,-6.69544,-7.44904,N.A.,7.03,-0.446363,214.218,15,4,-2.30926,0,-4.39571,-0.338125,-0.329295,0.20655,0.4704,0,0,TK_dhbt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-3,5,-6.22958,-7.38899,N.A.,7.03,-0.415305,214.218,15,4,-2.2431,0,-4.41978,-0.0248703,-0.367688,0.355463,0.4704,0,0,TK_dhbt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,0,-7.39418,-8.60088,N.A.,12.3,-0.410788,255.231,18,5,-2.76516,0,-4.865,-0.543599,-0.199843,0.391411,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,1,-7.55896,-8.76081,N.A.,12.3,-0.419942,255.231,18,5,-2.86486,0,-4.79188,-0.514753,-0.337239,0.361776,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,2,-7.46195,-8.70573,N.A.,12.3,-0.414553,255.231,18,5,-2.87589,0,-4.85565,-0.381672,-0.352157,0.415421,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,3,-7.17199,-8.19803,N.A.,12.3,-0.398444,255.231,18,5,-2.64917,0,-4.66113,-0.747237,-0.134428,0.431971,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,4,-7.01298,-8.59244,N.A.,12.3,-0.38961,255.231,18,5,-2.58262,0,-4.77562,-0.240042,-0.285319,0.282616,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,5,-7.52553,-8.15004,N.A.,12.3,-0.418085,255.231,18,5,-2.64966,0,-4.76986,-0.713593,-0.318898,0.338484,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,6,-7.31144,-8.26914,N.A.,12.3,-0.406191,255.231,18,5,-2.7626,0,-4.82037,-0.582827,-0.238989,0.505347,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,7,-7.41259,-8.83533,N.A.,12.3,-0.411811,255.231,18,5,-2.80861,0,-4.78761,-0.573004,-0.304457,0.473088,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,8,-6.79371,-8.32758,N.A.,12.3,-0.377428,255.231,18,5,-2.70353,0,-4.90304,-0.210429,-0.215563,0.650861,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,9,-7.07579,-7.9165,N.A.,12.3,-0.3931,255.231,18,5,-2.4304,0,-4.68891,-0.650243,-0.23311,0.338864,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-4,10,-7.16654,-8.34099,N.A.,12.3,-0.398141,255.231,18,5,-2.85169,0,-4.92061,-0.484601,-0.18955,0.691917,0.588,0,0,TK_ganciclovir,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-5,0,-4.65513,-9.44056,N.A.,7.42,-0.221673,296.279,21,3,-1.97625,0,-5.1372,0.937961,0.103589,1.06396,0.3528,0,0,TK_hmtt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-5,1,-3.75681,-9.6062,N.A.,7.42,-0.178895,296.279,21,3,-2.59478,0,-5.23811,0.936452,-0.224568,3.0114,0.3528,0,0,TK_hmtt,mnc32 11031721223D 1 1.00000 0.00000 0 tk3d.sdf-ligand-5,2,-3.4215,-12.1814,N.A.,7.42,-0.162928,296.279,21,3,-2.68148,0,-5.20993,-0.74364,-0.0364816,4.89723,0.3528,0,0,TK_hmtt,mnc32 11031721223D 1 1.00000 0.00000 0 |
该文件为CSV格式可以直接用Excel打开,进行进一步处理。比如,将每个分子Rank score为0的化合物过滤出来,考察VS score的虚拟筛选性能。本文上述ROC曲线就是用这个文件生成的。
如果您不想用gawk或R来处理,推荐用csvtk来对该文件进行排序等。比如,将每个化合物Rank score最佳的0那一行提取出来,也就是将第2列值为0的行过滤出来:
1 2 | bash命令: sed 1,4d decoy_score.csv |grep -v Non|csvtk filter -f "2=0" |
csvtk下载与安装:http://bioinf.shenwei.me/csvtk
三. 相关软件
- Flare
Flare与Lead Finder不同之处在于Flare主攻基于结构的设计: 分子对接(为结合模型预测而设计),水分子位置的预测,结合自由能计算,相互作用势分析,蛋白结构的结构准备、序列比对、分子叠合、等。而Lead Finder专注于虚拟筛选。
四. 相关文献
- Stroganov O.V. et al., Lead Finder: an approach to improve accuracy of protein-ligand docking, binding energy estimation, and virtual screening, J Chem Inf Model. 2008 Dec;48(12):2371-85. PubMed ID: 19007114
- Novikov F. et al., CSAR scoring challenge reveals the need for new concepts in estimating protein-ligand binding affinity, J Chem Inf Model. 2011 Sep 26;51(9):2090-2096. PubMed ID: 21612285
- Novikov F.N. et al., Improving performance of docking-based virtual screening by structural filtration, J Mol Model. 2010 Jul;16(7):1223-30. Epub 2009 Dec 30
- Novikov F.N. et al., Lead Finder docking and virtual screening evaluation with Astex and DUD test sets, J Comput Aided Mol Des. 2012 Jun;26(6):725-35
- Zeifman A.A. et al., Hit clustering can improve virtual fragment screening: CDK2 and PARP1 case studies, J Mol Model. 2012 Jun;18(6):2553-66. Epub 2011 Nov 9
- Stroganov O.V. et al., TSAR, a new graph-theoretical approach to computational modeling of protein side-chain flexibility: Modeling of ionization properties of proteins. Proteins, 2011 Sep; 79(9):2693-2710. PubMed ID: 21769942
五. 相关主题
- Flare 教程 | 分子对接-结合模式预测
- 十种分子对接软件的性能评估
- LigandScout教程–VINA分子对接
- 如何进行虚拟筛选的方法学验证
- 分子对接教程–用FRED进行虚拟筛选
http://blog.molcalx.com.cn/2017/06/04/flare-tutorial-dock-imatinib.html
http://blog.molcalx.com.cn/2017/02/08/evaluation-ten-docking-program.html
http://blog.molcalx.com.cn/2016/04/04/ligandscout-vina-tutorial.html
http://blog.molcalx.com.cn/2016/09/22/virtual-screening-methodology-validation.html
http://blog.molcalx.com.cn/2016/09/15/openeye-tutorial-fred.html
六. 文献
- Mehler, E. L. Self-Consistent, Free Energy Based Approximation To Calculate PH Dependent Electrostatic Effects in Proteins. J. Phys. Chem. 1996, 100 (39), 16006–16018.
- Mehler, E. L.; Guarnieri, F. A Self-Consistent, Microenvironment Modulated Screened Coulomb Potential Approximation to Calculate PH-Dependent Electrostatic Effects in Proteins. Biophys. J. 1999, 77 (1), 3–22.



